Introduction to tutorial 8
In this tutorial we propose an example concerning the usage of the Discrete Empirical Interpolation Method (implemented in the HyperReduction class) for the approximation of a non-linear function. The following image illustrates the computational domain used to discretize the problem
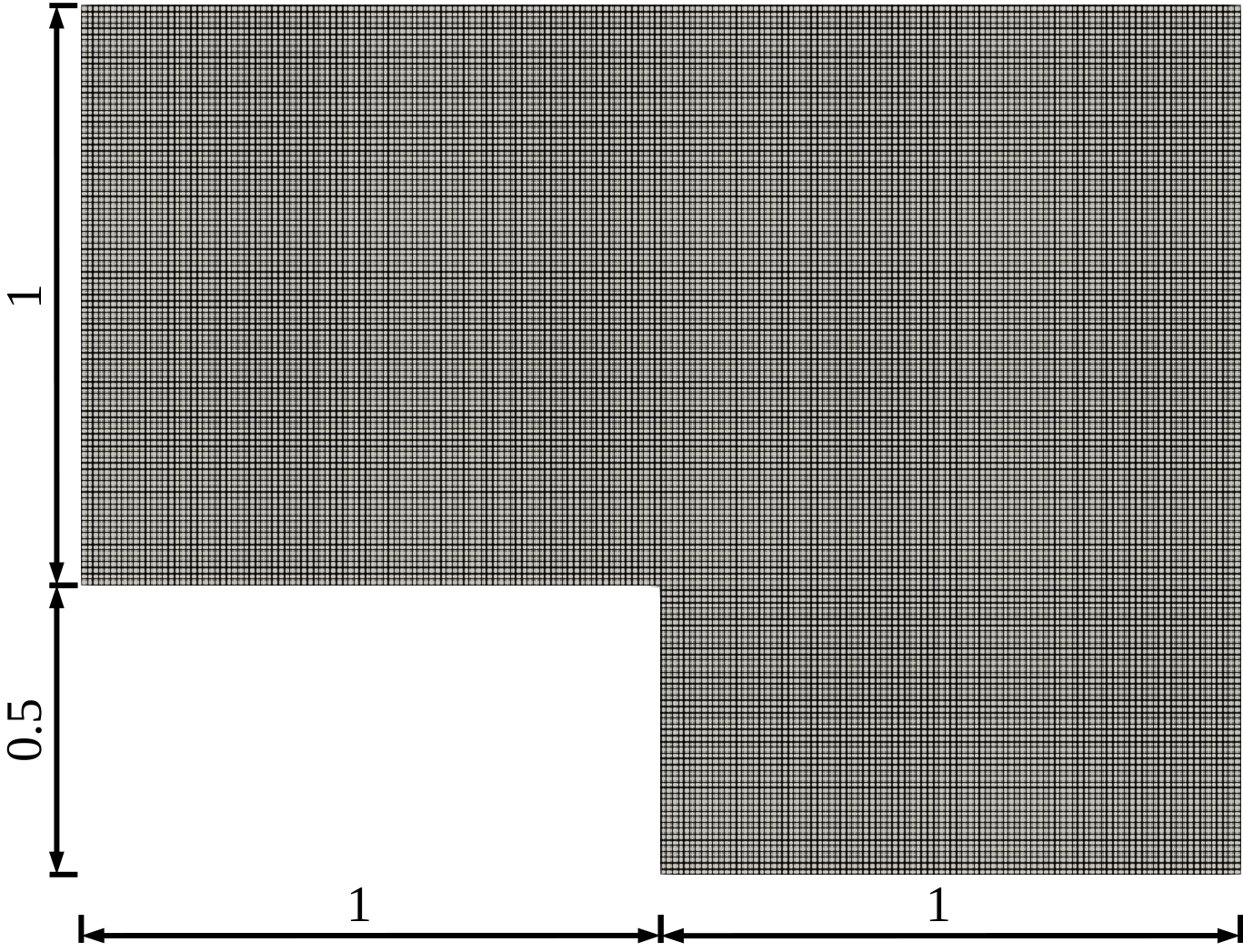
Computational Domain
The non-linear function is described by a parametric Gaussian function:
\[S(\mathbf{x},\mathbf{\mu}) = e^{-2(x-\mu_x-1)^2 - 2(y-\mu_y-0.5)^2},
\]
that is depending on the parameter vector \(\mathbf{\mu} = [\mu_x, \mu_y] \) and on the location inside the domain \( \mathbf{x} = [x,y] \). A training set of 100 samples is used to construct the "DEIM" modes which are later used for the approximation.
The necessary header files
First of all let's have a look to the header files that needs to be included and what they are responsible for: The header files of ITHACA-FV necessary for this tutorial are: <Foam2Eigen.H> for Eigen to OpenFOAM conversion of objects, <ITHACAstream.H> for ITHACA-FV input-output operations. <ITHACAPOD.H> for the POD decomposition, <hyperReduction.templates.H> for the "DEIM" approximation.
A detailed look into the code
The OpenFOAM header files:
#include "fvCFD.H"
#include "fvOptions.H"
#include "simpleControl.H"
#include "simpleControl.H"
#include "fvMeshSubset.H"
ITHACA-FV header files
Header file of the Foam2Eigen class.
Header file of the ITHACAPOD class.
Header file of the ITHACAstream class, it contains the implementation of several methods for input ou...
Header file of the ITHACAutilities namespace.
chrono to compute the speedup
Construction of the function to be approximated with the "DEIM" method from the HyperReduction class.
{
public:
HyperReduction(label n_modes, label n_nodes, Eigen::VectorXi initialSeeds, word problemName, SnapshotsLists &&...snapshotsLists)
Construct HyperReduction class, interpolation-based.
Method with the explicit definition of the non-linear function
static volScalarField evaluate_expression(volScalarField&
S, Eigen::MatrixXd mu)
{
volScalarField
yPos =
S.mesh().C().component(vector::Y).ref();
volScalarField
xPos =
S.mesh().C().component(vector::X).ref();
for (
auto i = 0;
i <
S.size();
i++)
{
S[
i] = std::exp(- 2 * std::pow(
xPos[
i] - mu(0) - 1,
2) - 2 * std::pow(
yPos[
i] - mu(1) - 0.5, 2));
}
volScalarField S(IOobject("S", runTime.timeName(), mesh, IOobject::NO_READ, IOobject::AUTO_WRITE), T.mesh(), dimensionedScalar("zero", dimensionSet(0, 0, -1, 1, 0, 0, 0), 0))
}
Method with the expression for the evaluation of the online coefficients
Eigen::VectorXd onlineCoeffs(Eigen::MatrixXd mu)
{
theta.resize(nodePoints().size());
auto f = evaluate_expression(subField(), mu);
for (
int i = 0;
i < nodePoints().size();
i++)
{
theta(
i) = f[localNodePoints[
i]];
}
return theta;
}
Now let's have a look to important command in the main function. Read parameters from the ITHACAdict file
int NDEIM =
para->ITHACAdict->lookupOrDefault<
int>(
"NDEIM", 15);
Class for the definition of some general parameters, the parameters must be defined from the file ITH...
static ITHACAparameters * getInstance()
Gets an instance of ITHACAparameters, to be used if the instance is already existing.
Definition of the parameter samples to train the non-linear function
Eigen::MatrixXd rand(label rows, label cols, double min, double max)
Generates random matrix with random values in an interval.
Offline training of the function and assembly of the snapshots list
for (
int i = 0;
i < 100;
i++)
{
}
static volScalarField evaluate_expression(volScalarField &S, Eigen::MatrixXd mu)
void exportSolution(GeometricField< Type, PatchField, GeoMesh > &s, fileName subfolder, fileName folder, word fieldName)
Export a field to file in a certain folder and subfolder.
Compute the "DEIM" modes using the ITHACAPOD class
Sp[0]).array();
static Eigen::MatrixXd PtrList2Eigen(PtrList< GeometricField< Type, PatchField, GeoMesh > > &fields, label Nfields=-1)
Convert a PtrList of snapshots to Eigen matrix (only internal field)
std::tuple< List< Eigen::SparseMatrix< double > >, List< Eigen::VectorXd > > DEIMmodes(List< Eigen::SparseMatrix< double > > &A, List< Eigen::VectorXd > &b, label nmodesA, label nmodesB, word MatrixName)
Get the DEIM modes for a generic non a parametrized matrix coming from a differential operator functi...
Eigen::VectorXd getMassMatrixFV(GeometricField< Type, PatchField, GeoMesh > &snapshot)
Gets a vector containing the volumes of each cell of the mesh.
Construction of the HyperReduction object passing the maximum number of "DEIM" modes NDEIM, the initSeeds (none in this tutorial), the name used to store the output "DEIM" and the list of snapshots Sp. The constructor is taking the number of modes and the number of point to compute, for the "DEIM" method these numbers are equal.
Eigen::VectorXi initSeeds(0);
Compute the "DEIM" method HyperReduction::offlineGappyDEIM passing the "DEIM" modes and the normalizingWeights
c.offlineGappyDEIM(snapshotsModes, normalizingWeights);
Command to generate the submeshes used for pointwise evaluation of the function
c.generateSubmesh(2,
mesh);
c.subField = c.interpolateField<volScalarField>(Sp[0]);
Definition of a new sample value to test the accuracy of the method \( \mu* = (0,0) \)
Eigen::MatrixXd par_new(2, 1);
par_new(0, 0) = 0;
par_new(1, 0) = 0;
Evaluation of the function using "DEIM" and reconstruction of the field
Eigen::VectorXd aprfield = c.MatrixOnline * c.onlineCoeffs(par_new);
static GeometricField< scalar, PatchField, GeoMesh > Eigen2field(GeometricField< scalar, PatchField, GeoMesh > &field, Eigen::VectorXd &eigen_vector, bool correctBC=true)
Convert a vector in Eigen format into an OpenFOAM scalar GeometricField.
Export the approximation
Evaluate the function on in \(\mu*\) using the FOM
Export the FOM field
Compute the error and print it
double errorL2Rel(GeometricField< T, fvPatchField, volMesh > &field1, GeometricField< T, fvPatchField, volMesh > &field2, List< label > *labels)
Computes the relative error between two geometric Fields in L2 norm.
The plain program
Here there's the plain code
#include "fvCFD.H"
#include "fvOptions.H"
#include "simpleControl.H"
#include "simpleControl.H"
#include "fvMeshSubset.H"
#include <chrono>
{
public:
{
volScalarField
yPos =
S.mesh().C().component(vector::Y).ref();
volScalarField
xPos =
S.mesh().C().component(vector::X).ref();
for (
auto i = 0;
i <
S.size();
i++)
{
S[
i] = std::exp(- 2 * std::pow(
xPos[
i] - mu(0) - 1,
2) - 2 * std::pow(
yPos[
i] - mu(1) - 0.5, 2));
}
}
{
{
}
}
PtrList<volScalarField>
fields;
};
int main(
int argc,
char* argv[])
{
#include "setRootCase.H"
Foam::Time
runTime(Foam::Time::controlDictName, args);
(
Foam::IOobject
(
Foam::fvMesh::defaultRegion,
Foam::IOobject::MUST_READ
)
);
int NDEIM =
para->ITHACAdict->lookupOrDefault<
int>(
"NDEIM", 15);
#include "createFields.H"
PtrList<volScalarField> Sp;
(
IOobject
(
"S",
IOobject::NO_READ,
IOobject::AUTO_WRITE
),
dimensionedScalar("zero", dimensionSet(0, 0, -1, 1, 0, 0, 0), 0)
);
for (
int i = 0;
i < 100;
i++)
{
}
Eigen::MatrixXd snapshotsModes;
Sp[0]).array();
Eigen::VectorXi initSeeds(0);
c.offlineGappyDEIM(snapshotsModes, normalizingWeights);
c.generateSubmesh(2,
mesh);
c.subField = c.interpolateField<volScalarField>(Sp[0]);
Eigen::MatrixXd par_new(2, 1);
par_new(0, 0) = 0;
par_new(1, 0) = 0;
Eigen::VectorXd aprfield = c.MatrixOnline * c.onlineCoeffs(par_new);
return 0;
}
autoPtr< volScalarField > subField
PtrList< volScalarField > fields
List< label > localNodePoints
autoPtr< IOList< label > > nodePoints
simpleControl simple(mesh)

 1.13.2
1.13.2